Overview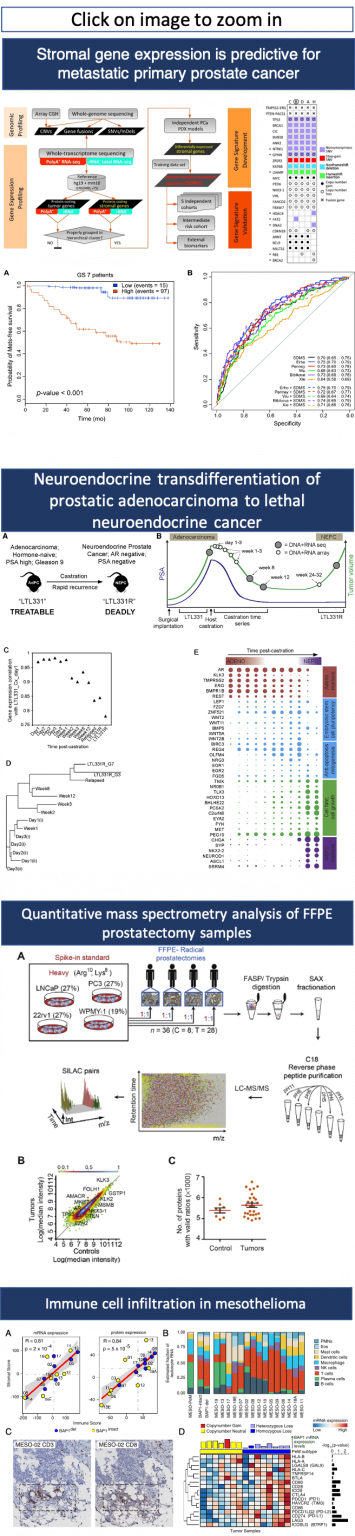
Dr. Collins has a B.Sc. in Biological Sciences from Western New England College, Springfield, MA, and a Ph.D. in Medical Genetics from the University of British Columbia. As a pioneer in translational genomics, Dr. Collins was deeply involved in the development of molecular cytogenetics1,2,3,4. He was the first to establish that genome copy number alteration predicts the risk of metastasis in prostate cancer patients5 and pioneered paired-end sequencing and its application to genome mapping strategies6. More recently, Dr. Collins has helped elucidate the mechanisms of transdifferentiation from prostatic adenocarcinoma to lethal neuroendocrine cancer7,8,9,10,11,12, which led to the development of liquid biopsies for prostate cancer13,14,15. He established that the tumour microenvironment participates in driving prostate cancer metastasis16,17 and identified, for the first time, therapeutic vulnerabilities of mesothelioma18,19.
Research Areas
Dr. Collins’ research focus is translational cancer genomics and precision oncology. The research focuses on four broad areas. The first area investigates how the tumour microenvironment promotes metastasis in advanced adenocarcinoma of the prostate. Dr. Collins’ team discovered that genes expressed in the tumour microenvironment are a strong predictor of metastasis16,17. The research includes patient-derived xenografts, single-cell sequencing, expression of microenvironment-specific long non-coding RNAs, and mass spectrometry proteomics. The second area is neuroendocrine transdifferentiation (NEtD) of prostatic adenocarcinoma to lethal neuroendocrine cancer. There is evidence that neuroendocrine prostate cancer may be more prevalent in Asia than in the western hemisphere. New potent androgen deprivation therapies appear to drive a switch or phase transition to a neuronal cell type. Dr. Collins’ laboratory was an early pioneer in the study of the mechanism driving NEtD 7,8,9,10,11,12. Preliminary new work suggests that a long non-coding RNA may be the integral switch. Single-cell RNA sequencing and proteomics are employed to understand this adaptive switch along with functional studies aimed at chromatin remodelling. The third area is tumour dormancy, a central problem in the management of cancer treatment when cancer cells evade therapy by entering a dormant state. In prostate cancer, disseminated dormant cells are generally found in the bone marrow. This makes their study extremely difficult and in vitro models do not recapitulate the complexity of dormancy. Dr. Collins’ studies have identified activation of an intrinsic dormancy pathway in response to androgen deprivation in high-fidelity patient-derived xenograft tumors21. Nematodes including the model organism C. elegans enters dormancy22 upon environmental stress. CRISPR-mediated deletion of relevant genes in C. elegans reduces the induction of dormancy in response to environmental stress. In addition, extrinsic microenvironment genes were found activated using a novel genomics approach only possible in PDX models16. The intrinsic and extrinsic genes are currently being pursued as potential drug targets. Fourth, Dr. Collins has an active research program into liquid biopsies for and therapeutic vulnerabilities of mesothelioma. Prostate cancer is a primary but not exclusive focus (refer to 18,19,20 for examples of non-prostate cancer research).
Dr. Collins has a deep interest in precision oncology23,24,25,26 and will collaborate broadly with interested oncologists and surgeons to improve outcomes.
Graduate and Postdoctoral Studies
Graduate students and postdoctoral fellows have the option to be co-mentored by Dr. Collins and a computer scientist if their primary interests lie in computer science. This setting allows cross-training in both cancer genomics and bioinformatics. Students may be based at the University of British Columbia or Simon Fraser University. Trainees have the option to carry out part of their research at local companies if research interests align. There may also be opportunities for trainees to spend time at Taipei Medical University in Taiwan or the Institute of Systems Genetics at West China Hospital and Sichuan University.
Alumni of the Collins’ laboratory have taken positions at institutions including UCSF, UCD, Yale, UBC, Harvard, Johns Hopkins, Joint Genome Institute, Weill Cornell Medical School, The National Cancer Institute, Kyoto University, Tianjin Medical University, Zheijiang University and throughout the private sector.
⇒ See the list of degree programs available with Dr. Collins.
References listed above
- Collins, C. et al. Construction and characterization of plasmid libraries enriched in sequences from single human chromosomes. Genomics 11, 997–1006 (1991).
- Collins, C. et al. A sequence-based survey of the complex structural organization of tumor genomes. Genome Biol. 9, 1 (2008).
- Raphael, B. J., Volik, S., Collins, C. & Pevzner, P. A. Reconstructing tumor genome architectures. Bioinformatics 19, ii162–ii171 (2003).
- Bashir, A., Volik, S., Collins, C., Bafna, V. & Raphael, B. J. Evaluation of paired-end sequencing strategies for detection of genome rearrangements in cancer. PLoS Comput Biol 4, e1000051 (2008).
- Paris, P. L. et al. A group of genome-based biomarkers that add to a Kattan nomogram for predicting progression in men with high-risk prostate cancer. Clin. Cancer Res. 16, 195–202 (2010).
- Volik, S. V et al. End sequence profiling (ESP): A sequence-based approach to structural analysis of tumor genomes. Cancer Res. 64, 1304 (2004).
- Lapuk, A. V et al. From sequence to molecular pathology, and a mechanism driving the neuroendocrine phenotype in prostate cancer. J. Pathol. 227, 286–297 (2012).
- Lin, D. et al. High fidelity patient-derived xenografts for accelerating prostate cancer discovery and drug development. Cancer Res. 74, 1272–1283 (2014).
- Akamatsu, S. et al. The placental gene PEG10 promotes progression of neuroendocrine prostate cancer. Cell Rep. 12, 922–936 (2015).
- Li, Y. et al. SRRM4 Drives Neuroendocrine Transdifferentiation of Prostate Adenocarcinoma Under Androgen Receptor Pathway Inhibition. Eur. Urol. 71, 68–78 (2017).
- Li, Y. et al. RNA Splicing of the BHC80 Gene Contributes to Neuroendocrine Prostate Cancer Progression. Eur. Urol. 1–10 (2019). doi:10.1016/j.eururo.2019.03.011
- Flores-morales, A. et al. Proteogenomic Characterization of Patient- Derived Xenografts Highlights the Role of REST in Neuroendocrine Differentiation of Castration- Resistant Prostate Cancer. (2019). doi:10.1158/1078-0432.CCR-18-0729
- Azad, A. A. et al. Androgen receptor gene aberrations in circulating cell-free DNA: Biomarkers of therapeutic resistance in castration-resistant prostate cancer. Clin. Cancer Res. 21, 2315–2324 (2015).
- Lallous, N. et al. Functional analysis of androgen receptor mutations that confer anti-androgen resistance identified in circulating cell-free DNA from prostate cancer patients. Genome Biol. 17, 1 (2016).
- Wyatt, A. W. et al. Genomic Alterations in Cell-Free DNA and Enzalutamide Resistance in Castration-Resistant Prostate Cancer. (2019). doi:10.1001/jamaoncol.2016.0494
- Mo, F. et al. Stromal Gene Expression is Predictive for Metastatic Primary Prostate Cancer § Associate Editor : Eur. Urol. 1–9 (2017). doi:10.1016/j.eururo.2017.02.038
- Mo, F. et al. Re: Stromal Gene Expression is Predictive for Metastatic Primary Prostate Cancer. Eur. Urol. 73, 478 (2018).
- Shrestha, R. et al. BAP1 haploinsufficiency predicts a distinct immunogenic class of malignant peritoneal mesothelioma. 1–12 (2019).
- Ladanyi, M., Sanchez Vega, F. & Zauderer, M. Loss of BAP1 as a candidate predictive biomarker for immunotherapy of mesothelioma. Genome Med. 11, 10–12 (2019).
- Fernandez, M. L. et al. Markers of MEK inhibitor resistance in low-grade serous ovarian cancer: EGFR is a potential therapeutic target. Cancer Cell Int. 19, 1–17 (2019).
- Lin, D. et al. High fidelity patient-derived xenografts for accelerating prostate cancer discovery and drug development. Cancer Res. 74, 1272–1283 (2014).
- Fielenbach, N. & Antebi, A. C. elegans dauer formation and the molecular basis of plasticity. Genes Dev. 22, 2149–65 (2008).
- Hodzic, E. et al. Combinatorial Detection of Conserved Alteration Patterns for Identifying Cancer Subnetworks. 1–13 (2019). doi:10.1093/gigascience/giz024
- Sharifi-Noghabi, H., Zolotareva, O., Collins, C. C. & Ester, M. MOLI: Multi-Omics Late Integration with deep neural networks for drug response prediction. bioRxiv 531327 (2019). doi:10.1101/531327
- Sharifi-Noghabi, H. et al. Deep Genomic Signature for early metastasis prediction in prostate cancer. bioRxiv 276055 (2019). doi:10.1101/276055
- Shrestha, R., Hodzic, E., Sauerwald, T. & Dao, P. HIT ’ nDRIVE : Patient-Specific Multi-Driver Gene Prioritization for Precision Oncology. 1573–1588 doi:10.1101/gr.221218.117.10